What would happen to cell if they did not differentiate

Stalk cell differentiation into various tissue types.

Prison cell-count distribution featuring cellular differentiation for three types of cells (progenitor , osteoblast , and chondrocyte ) exposed to pro-osteoblast stimulus.[ane]
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one[2] [3] Commonly, the jail cell changes to a more specialized blazon. Differentiation happens multiple times during the development of a multicellular organism every bit it changes from a unproblematic zygote to a circuitous system of tissues and prison cell types. Differentiation continues in adulthood every bit adult stem cells divide and create fully differentiated daughter cells during tissue repair and during normal cell turnover. Some differentiation occurs in response to antigen exposure. Differentiation dramatically changes a prison cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals. These changes are largely due to highly controlled modifications in factor expression and are the study of epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the Dna sequence itself. Although metabolic composition does get altered quite dramatically[4] where stem cells are characterized past arable metabolites with highly unsaturated structures whose levels decrease upon differentiation. Thus, different cells can have very different concrete characteristics despite having the aforementioned genome.
A specialized blazon of differentiation, known as final differentiation, is of importance in some tissues, for case vertebrate nervous organisation, striated musculus, epidermis and gut. During terminal differentiation, a precursor cell formerly capable of cell division, permanently leaves the cell cycle, dismantles the prison cell cycle machinery and frequently expresses a range of genes characteristic of the cell'south concluding office (e.g. myosin and actin for a muscle jail cell). Differentiation may continue to occur after terminal differentiation if the capacity and functions of the cell undergo further changes.
Amongst dividing cells, there are multiple levels of cell authority, the prison cell's ability to differentiate into other prison cell types. A greater authorization indicates a larger number of cell types that can be derived. A cell that tin can differentiate into all cell types, including the placental tissue, is known equally totipotent. In mammals, only the zygote and subsequent blastomeres are totipotent, while in plants, many differentiated cells tin can go totipotent with simple laboratory technique. A cell that can differentiate into all cell types of the adult organism is known as pluripotent. Such cells are called meristematic cells in higher plants and embryonic stem cells in animals, though some groups report the presence of developed pluripotent cells. Virally induced expression of iv transcription factors Oct4, Sox2, c-Myc, and Klf4 (Yamanaka factors) is sufficient to create pluripotent (iPS) cells from adult fibroblasts.[5] A multipotent cell is one that can differentiate into multiple different, but closely related cell types.[half-dozen] Oligopotent cells are more restricted than multipotent, but can still differentiate into a few closely related cell types.[6] Finally, unipotent cells can differentiate into only ane cell blazon, but are capable of self-renewal.[6] In cytopathology, the level of cellular differentiation is used equally a measure out of cancer progression. "Form" is a marker of how differentiated a prison cell in a tumor is.[7]
Mammalian jail cell types [edit]
Iii bones categories of cells make up the mammalian body: germ cells, somatic cells, and stem cells. Each of the approximately 37.2 trillion (3.72x10thirteen) cells in an developed human has its own copy or copies of the genome except certain cell types, such as ruby-red claret cells, that lack nuclei in their fully differentiated country. Nigh cells are diploid; they have two copies of each chromosome. Such cells, called somatic cells, make upwardly most of the human body, such as pare and musculus cells. Cells differentiate to specialize for dissimilar functions.[8]
Germ line cells are whatsoever line of cells that give rising to gametes—eggs and sperm—and thus are continuous through the generations. Stem cells, on the other hand, have the ability to divide for indefinite periods and to requite rise to specialized cells. They are best described in the context of normal man development.[ citation needed ]
Development begins when a sperm fertilizes an egg and creates a single jail cell that has the potential to form an unabridged organism. In the starting time hours after fertilization, this jail cell divides into identical cells. In humans, approximately four days later fertilization and after several cycles of jail cell partition, these cells begin to specialize, forming a hollow sphere of cells, called a blastocyst.[9] The blastocyst has an outer layer of cells, and inside this hollow sphere, there is a cluster of cells called the inner cell mass. The cells of the inner jail cell mass go on to form virtually all of the tissues of the man body. Although the cells of the inner jail cell mass tin course nearly every type of jail cell found in the man body, they cannot form an organism. These cells are referred to as pluripotent.[ten]
Pluripotent stem cells undergo farther specialization into multipotent progenitor cells that then give rise to functional cells. Examples of stalk and progenitor cells include:[ citation needed ]
- Radial glial cells (embryonic neural stem cells) that give ascent to excitatory neurons in the fetal encephalon through the process of neurogenesis.[eleven] [12] [13]
- Hematopoietic stalk cells (adult stem cells) from the bone marrow that give rise to red blood cells, white claret cells, and platelets
- Mesenchymal stalk cells (developed stem cells) from the os marrow that give rise to stromal cells, fat cells, and types of bone cells
- Epithelial stem cells (progenitor cells) that give rise to the diverse types of pare cells
- Muscle satellite cells (progenitor cells) that contribute to differentiated muscle tissue.
A pathway that is guided by the cell adhesion molecules consisting of four amino acids, arginine, glycine, asparagine, and serine, is created as the cellular blastomere differentiates from the single-layered blastula to the three primary layers of germ cells in mammals, namely the ectoderm, mesoderm and endoderm (listed from virtually distal (exterior) to proximal (interior)). The ectoderm ends up forming the peel and the nervous organisation, the mesoderm forms the bones and muscular tissue, and the endoderm forms the internal organ tissues.
Dedifferentiation [edit]

Micrograph of a liposarcoma with some dedifferentiation, that is not identifiable as a liposarcoma, (left edge of image) and a differentiated component (with lipoblasts and increased vascularity (right of image)). Fully differentiated (morphologically beneficial) adipose tissue (middle of the epitome) has few blood vessels. H&Eastward stain.
Dedifferentiation, or integration, is a cellular process often seen in more basal life forms such equally worms and amphibians in which a partially or terminally differentiated jail cell reverts to an earlier developmental stage, usually as role of a regenerative process.[14] [15] Dedifferentiation also occurs in plants.[16] Cells in cell culture can lose properties they originally had, such as protein expression, or modify shape. This process is also termed dedifferentiation.[17]
Some believe dedifferentiation is an aberration of the normal evolution bike that results in cancer,[18] whereas others believe it to exist a natural office of the allowed response lost by humans at some point as a effect of evolution.
A modest molecule dubbed reversine, a purine analog, has been discovered that has proven to induce dedifferentiation in myotubes. These dedifferentiated cells could then redifferentiate into osteoblasts and adipocytes.[19]

Diagram exposing several methods used to revert adult somatic cells to totipotency or pluripotency.
Mechanisms [edit]

Mechanisms of cellular differentiation.
Each specialized cell type in an organism expresses a subset of all the genes that constitute the genome of that species. Each cell type is defined by its particular design of regulated gene expression. Jail cell differentiation is thus a transition of a prison cell from ane cell type to another and information technology involves a switch from 1 pattern of gene expression to another. Cellular differentiation during development tin can be understood as the effect of a gene regulatory network. A regulatory gene and its cis-regulatory modules are nodes in a gene regulatory network; they receive input and create output elsewhere in the network.[20] The systems biological science approach to developmental biology emphasizes the importance of investigating how developmental mechanisms collaborate to produce anticipated patterns (morphogenesis). All the same, an alternative view has been proposed recently[ when? ] [ by whom? ]. Based on stochastic gene expression, cellular differentiation is the result of a Darwinian selective process occurring amidst cells. In this frame, protein and gene networks are the result of cellular processes and not their cause.[ citation needed ]
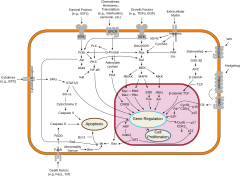
An overview of major indicate transduction pathways.
While evolutionarily conserved molecular processes are involved in the cellular mechanisms underlying these switches, in creature species these are very different from the well-characterized factor regulatory mechanisms of bacteria, and even from those of the animals' closest unicellular relatives.[21] Specifically, cell differentiation in animals is highly dependent on biomolecular condensates of regulatory proteins and enhancer Deoxyribonucleic acid sequences.
Cellular differentiation is oft controlled by jail cell signaling. Many of the signal molecules that convey information from prison cell to cell during the control of cellular differentiation are called growth factors. Although the details of specific betoken transduction pathways vary, these pathways often share the post-obit full general steps. A ligand produced by one cell binds to a receptor in the extracellular region of another cell, inducing a conformational alter in the receptor. The shape of the cytoplasmic domain of the receptor changes, and the receptor acquires enzymatic activeness. The receptor so catalyzes reactions that phosphorylate other proteins, activating them. A pour of phosphorylation reactions somewhen activates a dormant transcription factor or cytoskeletal protein, thus contributing to the differentiation procedure in the target jail cell.[22] Cells and tissues can vary in competence, their power to respond to external signals.[23]
Signal induction refers to cascades of signaling events, during which a jail cell or tissue signals to another jail cell or tissue to influence its developmental fate.[23] Yamamoto and Jeffery[24] investigated the role of the lens in middle formation in cave- and surface-dwelling house fish, a hit example of induction.[23] Through reciprocal transplants, Yamamoto and Jeffery[24] found that the lens vesicle of surface fish tin induce other parts of the heart to develop in cave- and surface-dwelling fish, while the lens vesicle of the cave-dwelling fish cannot.[23]
Other of import mechanisms fall under the category of asymmetric cell divisions, divisions that requite ascension to daughter cells with distinct developmental fates. Asymmetric cell divisions tin occur considering of asymmetrically expressed maternal cytoplasmic determinants or because of signaling.[23] In the old machinery, distinct girl cells are created during cytokinesis because of an uneven distribution of regulatory molecules in the parent jail cell; the singled-out cytoplasm that each daughter jail cell inherits results in a distinct design of differentiation for each daughter cell. A well-studied example of pattern formation by asymmetric divisions is body axis patterning in Drosophila. RNA molecules are an of import type of intracellular differentiation control indicate. The molecular and genetic basis of disproportionate cell divisions has also been studied in greenish algae of the genus Volvox, a model system for studying how unicellular organisms can evolve into multicellular organisms.[23] In Volvox carteri, the 16 cells in the inductive hemisphere of a 32-cell embryo divide asymmetrically, each producing one large and one pocket-sized girl cell. The size of the jail cell at the end of all cell divisions determines whether it becomes a specialized germ or somatic cell.[23] [25]
Epigenetic control [edit]
Since each cell, regardless of cell blazon, possesses the same genome, determination of cell type must occur at the level of gene expression. While the regulation of cistron expression can occur through cis- and trans-regulatory elements including a gene'southward promoter and enhancers, the trouble arises as to how this expression pattern is maintained over numerous generations of cell partition. Every bit it turns out, epigenetic processes play a crucial role in regulating the decision to adopt a stem, progenitor, or mature cell fate. This department will focus primarily on mammalian stem cells.
In systems biology and mathematical modeling of factor regulatory networks, jail cell-fate conclusion is predicted to showroom certain dynamics, such as attractor-convergence (the attractor can be an equilibrium point, limit cycle or foreign attractor) or oscillatory.[26]
Importance of epigenetic control [edit]
The offset question that tin exist asked is the extent and complexity of the role of epigenetic processes in the decision of cell fate. A clear reply to this question tin can be seen in the 2011 paper past Lister R, et al. [27] on aberrant epigenomic programming in human induced pluripotent stem cells. Equally induced pluripotent stem cells (iPSCs) are thought to mimic embryonic stem cells in their pluripotent properties, few epigenetic differences should exist between them. To test this prediction, the authors conducted whole-genome profiling of Dna methylation patterns in several human being embryonic stalk cell (ESC), iPSC, and progenitor cell lines.
Female adipose cells, lung fibroblasts, and foreskin fibroblasts were reprogrammed into induced pluripotent state with the OCT4, SOX2, KLF4, and MYC genes. Patterns of DNA methylation in ESCs, iPSCs, somatic cells were compared. Lister R, et al. observed significant resemblance in methylation levels between embryonic and induced pluripotent cells. Around eighty% of CG dinucleotides in ESCs and iPSCs were methylated, the same was true of simply 60% of CG dinucleotides in somatic cells. In add-on, somatic cells possessed minimal levels of cytosine methylation in non-CG dinucleotides, while induced pluripotent cells possessed like levels of methylation equally embryonic stem cells, between 0.5 and 1.5%. Thus, consistent with their corresponding transcriptional activities,[27] Dna methylation patterns, at least on the genomic level, are similar betwixt ESCs and iPSCs.
Nonetheless, upon examining methylation patterns more closely, the authors discovered 1175 regions of differential CG dinucleotide methylation betwixt at to the lowest degree one ES or iPS jail cell line. By comparing these regions of differential methylation with regions of cytosine methylation in the original somatic cells, 44-49% of differentially methylated regions reflected methylation patterns of the respective progenitor somatic cells, while 51-56% of these regions were unlike to both the progenitor and embryonic cell lines. In vitro-induced differentiation of iPSC lines saw transmission of 88% and 46% of hyper and hypo-methylated differentially methylated regions, respectively.
Two conclusions are readily apparent from this study. First, epigenetic processes are heavily involved in cell fate determination, every bit seen from the like levels of cytosine methylation betwixt induced pluripotent and embryonic stem cells, consistent with their respective patterns of transcription. Second, the mechanisms of reprogramming (and past extension, differentiation) are very complex and cannot exist easily duplicated, equally seen by the significant number of differentially methylated regions between ES and iPS prison cell lines. At present that these two points have been established, we tin examine some of the epigenetic mechanisms that are thought to regulate cellular differentiation.
Mechanisms of epigenetic regulation [edit]
Pioneer factors (Oct4, Sox2, Nanog) [edit]
Three transcription factors, OCT4, SOX2, and NANOG – the first two of which are used in induced pluripotent stem cell (iPSC) reprogramming, forth with Klf4 and c-Myc – are highly expressed in undifferentiated embryonic stem cells and are necessary for the maintenance of their pluripotency.[28] Information technology is thought that they attain this through alterations in chromatin construction, such as histone modification and Dna methylation, to restrict or permit the transcription of target genes. While highly expressed, their levels crave a precise residual to maintain pluripotency, perturbation of which volition promote differentiation towards different lineages based on how the gene expression levels modify. Differential regulation of October-4 and SOX2 levels take been shown to precede germ layer fate selection.[29] Increased levels of Oct4 and decreased levels of Sox2 promote a mesendodermal fate, with Oct4 actively suppressing genes associated with a neural ectodermal fate. Similarly, Increased levels of Sox2 and decreased levels of Oct4 promote differentiation towards a neural ectodermal fate, with Sox2 inhibiting differentiation towards a mesendodermal fate. Regardless of the lineage cells differentiate downwardly, suppression of NANOG has been identified as a necessary prerequisite for differentiation.[29]
Polycomb repressive complex (PRC2) [edit]
In the realm of cistron silencing, Polycomb repressive complex 2, one of two classes of the Polycomb group (PcG) family unit of proteins, catalyzes the di- and tri-methylation of histone H3 lysine 27 (H3K27me2/me3).[28] [30] [31] Past binding to the H3K27me2/iii-tagged nucleosome, PRC1 (also a complex of PcG family proteins) catalyzes the mono-ubiquitinylation of histone H2A at lysine 119 (H2AK119Ub1), blocking RNA polymerase Two action and resulting in transcriptional suppression.[28] PcG knockout ES cells do non differentiate efficiently into the three germ layers, and deletion of the PRC1 and PRC2 genes leads to increased expression of lineage-affiliated genes and unscheduled differentiation.[28] Presumably, PcG complexes are responsible for transcriptionally repressing differentiation and development-promoting genes.
Trithorax group proteins (TrxG) [edit]
Alternately, upon receiving differentiation signals, PcG proteins are recruited to promoters of pluripotency transcription factors. PcG-deficient ES cells tin begin differentiation just cannot maintain the differentiated phenotype.[28] Simultaneously, differentiation and development-promoting genes are activated past Trithorax group (TrxG) chromatin regulators and lose their repression.[28] [31] TrxG proteins are recruited at regions of loftier transcriptional action, where they catalyze the trimethylation of histone H3 lysine 4 (H3K4me3) and promote gene activation through histone acetylation.[31] PcG and TrxG complexes engage in straight contest and are thought to exist functionally antagonistic, creating at differentiation and development-promoting loci what is termed a "bivalent domain" and rendering these genes sensitive to rapid induction or repression.[32]
DNA methylation [edit]
Regulation of gene expression is farther accomplished through DNA methylation, in which the DNA methyltransferase-mediated methylation of cytosine residues in CpG dinucleotides maintains heritable repression past decision-making Deoxyribonucleic acid accessibility.[32] The majority of CpG sites in embryonic stem cells are unmethylated and appear to be associated with H3K4me3-carrying nucleosomes.[28] Upon differentiation, a small number of genes, including OCT4 and NANOG,[32] are methylated and their promoters repressed to forestall their farther expression. Consistently, Deoxyribonucleic acid methylation-deficient embryonic stem cells speedily enter apoptosis upon in vitro differentiation.[28]
Nucleosome positioning [edit]
While the DNA sequence of about cells of an organism is the same, the binding patterns of transcription factors and the corresponding gene expression patterns are dissimilar. To a big extent, differences in transcription gene binding are determined by the chromatin accessibility of their binding sites through histone modification and/or pioneer factors. In particular, information technology is important to know whether a nucleosome is covering a given genomic binding site or not. This tin be determined using a chromatin immunoprecipitation (ChIP) assay.[33]
Histone acetylation and methylation [edit]
Deoxyribonucleic acid-nucleosome interactions are characterized by two states: either tightly bound by nucleosomes and transcriptionally inactive, chosen heterochromatin, or loosely bound and usually, simply not always, transcriptionally active, called euchromatin. The epigenetic processes of histone methylation and acetylation, and their inverses demethylation and deacetylation primarily account for these changes. The effects of acetylation and deacetylation are more predictable. An acetyl grouping is either added to or removed from the positively charged Lysine residues in histones past enzymes called histone acetyltransferases or histone deacteylases, respectively. The acetyl grouping prevents Lysine's association with the negatively charged Deoxyribonucleic acid backbone. Methylation is not as straightforward, every bit neither methylation nor demethylation consistently correlate with either cistron activation or repression. Nevertheless, certain methylations have been repeatedly shown to either activate or repress genes. The trimethylation of lysine 4 on histone iii (H3K4Me3) is associated with factor activation, whereas trimethylation of lysine 27 on histone iii represses genes[34] [35] [36]
In stem cells [edit]
During differentiation, stem cells change their gene expression profiles. Contempo studies have implicated a role for nucleosome positioning and histone modifications during this process.[37] There are ii components of this process: turning off the expression of embryonic stem cell (ESC) genes, and the activation of cell fate genes. Lysine specific demethylase one (KDM1A) is thought to forestall the employ of enhancer regions of pluripotency genes, thereby inhibiting their transcription.[38] Information technology interacts with Mi-2/NuRD complex (nucleosome remodelling and histone deacetylase) complex,[38] giving an instance where methylation and acetylation are not discrete and mutually exclusive, but intertwined processes.
Role of signaling in epigenetic command [edit]
A final question to ask concerns the role of prison cell signaling in influencing the epigenetic processes governing differentiation. Such a role should be, as it would be reasonable to recollect that extrinsic signaling tin can pb to epigenetic remodeling, only equally information technology can lead to changes in gene expression through the activation or repression of different transcription factors. Trivial direct data is available concerning the specific signals that influence the epigenome, and the bulk of current knowledge about the subject consists of speculations on plausible candidate regulators of epigenetic remodeling.[39] We will commencement discuss several major candidates thought to be involved in the induction and maintenance of both embryonic stem cells and their differentiated progeny, and and so turn to one instance of specific signaling pathways in which more direct evidence exists for its role in epigenetic change.
The first major candidate is Wnt signaling pathway. The Wnt pathway is involved in all stages of differentiation, and the ligand Wnt3a tin can substitute for the overexpression of c-Myc in the generation of induced pluripotent stem cells.[39] On the other hand, disruption of β-catenin, a component of the Wnt signaling pathway, leads to decreased proliferation of neural progenitors.
Growth factors contain the second major prepare of candidates of epigenetic regulators of cellular differentiation. These morphogens are crucial for development, and include bone morphogenetic proteins, transforming growth factors (TGFs), and fibroblast growth factors (FGFs). TGFs and FGFs accept been shown to sustain expression of OCT4, SOX2, and NANOG by downstream signaling to Smad proteins.[39] Depletion of growth factors promotes the differentiation of ESCs, while genes with bivalent chromatin can get either more restrictive or permissive in their transcription.[39]
Several other signaling pathways are as well considered to exist master candidates. Cytokine leukemia inhibitory factors are associated with the maintenance of mouse ESCs in an undifferentiated state. This is accomplished through its activation of the Jak-STAT3 pathway, which has been shown to exist necessary and sufficient towards maintaining mouse ESC pluripotency.[40] Retinoic acid can induce differentiation of human being and mouse ESCs,[39] and Notch signaling is involved in the proliferation and self-renewal of stem cells. Finally, Sonic hedgehog, in addition to its role equally a morphogen, promotes embryonic stem cell differentiation and the self-renewal of somatic stem cells.[39]
The problem, of course, is that the candidacy of these signaling pathways was inferred primarily on the basis of their role in evolution and cellular differentiation. While epigenetic regulation is necessary for driving cellular differentiation, they are certainly not sufficient for this process. Straight modulation of factor expression through modification of transcription factors plays a key role that must be distinguished from heritable epigenetic changes that tin can persist fifty-fifty in the absence of the original ecology signals. Only a few examples of signaling pathways leading to epigenetic changes that alter cell fate currently exist, and we will focus on one of them.
Expression of Shh (Sonic hedgehog) upregulates the production of BMI1, a component of the PcG circuitous that recognizes H3K27me3. This occurs in a Gli-dependent mode, as Gli1 and Gli2 are downstream effectors of the Hedgehog signaling pathway. In civilisation, Bmi1 mediates the Hedgehog pathway's ability to promote human being mammary stem cell cocky-renewal.[41] In both humans and mice, researchers showed Bmi1 to be highly expressed in proliferating immature cerebellar granule cell precursors. When Bmi1 was knocked out in mice, impaired cerebellar evolution resulted, leading to significant reductions in postnatal brain mass along with abnormalities in motor control and behavior.[42] A dissever study showed a significant decrease in neural stem cell proliferation along with increased astrocyte proliferation in Bmi cipher mice.[43]
An alternative model of cellular differentiation during embryogenesis is that positional information is based on mechanical signalling past the cytoskeleton using Embryonic differentiation waves. The mechanical signal is then epigenetically transduced via signal transduction systems (of which specific molecules such every bit Wnt are function) to consequence in differential gene expression.
In summary, the office of signaling in the epigenetic command of cell fate in mammals is largely unknown, but distinct examples exist that point the likely existence of further such mechanisms.
Effect of matrix elasticity [edit]
In guild to fulfill the purpose of regenerating a variety of tissues, adult stems are known to migrate from their niches, adhere to new extracellular matrices (ECM) and differentiate. The ductility of these microenvironments are unique to different tissue types. The ECM surrounding encephalon, muscle and bone tissues range from soft to stiff. The transduction of the stem cells into these cells types is not directed solely by chemokine cues and cell to prison cell signaling. The elasticity of the microenvironment can as well affect the differentiation of mesenchymal stem cells (MSCs which originate in bone marrow.) When MSCs are placed on substrates of the same stiffness equally encephalon, muscle and bone ECM, the MSCs have on properties of those respective cell types.[44] Matrix sensing requires the cell to pull confronting the matrix at focal adhesions, which triggers a cellular mechano-transducer to generate a signal to be informed what force is needed to deform the matrix. To determine the key players in matrix-elasticity-driven lineage specification in MSCs, unlike matrix microenvironments were mimicked. From these experiments, information technology was concluded that focal adhesions of the MSCs were the cellular mechano-transducer sensing the differences of the matrix elasticity. The non-musculus myosin IIa-c isoforms generates the forces in the prison cell that pb to signaling of early commitment markers. Nonmuscle myosin IIa generates the to the lowest degree force increasing to non-muscle myosin IIc. In that location are also factors in the prison cell that inhibit not-musculus myosin II, such as blebbistatin. This makes the jail cell effectively blind to the surrounding matrix.[44] Researchers have obtained some success in inducing stem prison cell-like backdrop in HEK 239 cells by providing a soft matrix without the use of diffusing factors.[45] The stem-jail cell backdrop appear to be linked to tension in the cells' actin network. One identified mechanism for matrix-induced differentiation is tension-induced proteins, which remodel chromatin in response to mechanical stretch.[46] The RhoA pathway is also implicated in this process.
Evolutionary history [edit]
A billion-years-sometime, likely holozoan, protist, Bicellum brasieri with ii types of cells, shows that the evolution of differentiated multicellularity, possibly merely not necessarily of animal lineages, occurred at least ane billion years ago and possibly mainly in freshwater lakes rather than the ocean.[47] [48] [49] [ description needed ]
See also [edit]
- Interbilayer Forces in Membrane Fusion
- Fusion mechanism
- Lipid bilayer fusion
- Cell-cell fusogens
- CAF-i
- Listing of human cell types derived from the germ layers
References [edit]
- ^ Kryven, I.; Röblitz, S.; Schütte, Ch. (2015). "Solution of the chemic main equation by radial ground functions approximation with interface tracking". BMC Systems Biology. 9 (1): 67. doi:10.1186/s12918-015-0210-y. PMC4599742. PMID 26449665.

- ^ Slack, J.One thousand.W. (2013) Essential Developmental Biology. Wiley-Blackwell, Oxford.
- ^ Slack, J.1000.W. (2007). "Metaplasia and transdifferentiation: from pure biology to the clinic". Nature Reviews Molecular Prison cell Biological science. 8 (five): 369–378. doi:10.1038/nrm2146. PMID 17377526. S2CID 3353748.
- ^ Yanes, Oscar; Clark, Julie; Wong, Diana 1000.; Patti, Gary J.; Sánchez-Ruiz, Antonio; Benton, H. Paul; Trauger, Sunia A.; Desponts, Caroline; Ding, Sheng; Siuzdak, Gary (June 2010). "Metabolic oxidation regulates embryonic stem cell differentiation". Nature Chemical Biology. 6 (6): 411–417. doi:10.1038/nchembio.364. ISSN 1552-4469. PMC2873061. PMID 20436487.
- ^ Takahashi, One thousand; Yamanaka, S (2006). "Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures past defined factors". Jail cell. 126 (four): 663–76. doi:10.1016/j.cell.2006.07.024. hdl:2433/159777. PMID 16904174. S2CID 1565219.
- ^ a b c Schöler, Hans R. (2007). "The Potential of Stem Cells: An Inventory". In Nikolaus Knoepffler; Dagmar Schipanski; Stefan Lorenz Sorgner (eds.). Humanbiotechnology as Social Claiming. Ashgate Publishing. p. 28. ISBN978-0-7546-5755-2.
- ^ "NCI Dictionary of Cancer Terms". National Cancer Institute. Retrieved 1 November 2013.
- ^ Lodish, Harvey (2000). Molecular Prison cell Biology (4th ed.). New York: Due west. H. Freeman. Section fourteen.ii. ISBN978-0-7167-3136-8.
- ^ Kumar, Rani (2008). Textbook of Human being Embryology. I.K. International Publishing House. p. 22. ISBN9788190675710.
- ^ D. Binder, Marc; Hirokawa, Nobutaka; Windhorst, Uwe (2009). Encyclopedia of Neuroscience. Springer. ISBN978-3540237358.
- ^ Rakic, P (October 2009). "Evolution of the neocortex: a perspective from developmental biology". Nature Reviews. Neuroscience. 10 (x): 724–35. doi:ten.1038/nrn2719. PMC2913577. PMID 19763105.
- ^ Lui, JH; Hansen, DV; Kriegstein, AR (8 July 2011). "Development and evolution of the human neocortex". Cell. 146 (1): 18–36. doi:ten.1016/j.cell.2011.06.030. PMC3610574. PMID 21729779.
- ^ Rash, BG; Ackman, JB; Rakic, P (February 2016). "Bidirectional radial Ca(2+) activity regulates neurogenesis and migration during early on cortical column germination". Science Advances. two (2): e1501733. Bibcode:2016SciA....2E1733R. doi:10.1126/sciadv.1501733. PMC4771444. PMID 26933693.
- ^ Stocum DL (2004). "Amphibian regeneration and stem cells". Curr. Top. Microbiol. Immunol. Current Topics in Microbiology and Immunology. 280: i–70. doi:10.1007/978-3-642-18846-6_1. ISBN978-3-540-02238-1. PMID 14594207.
- ^ Casimir CM, Gates Atomic number 82, Patient RK, Brockes JP (1988-12-01). "Evidence for dedifferentiation and metaplasia in amphibian limb regeneration from inheritance of Deoxyribonucleic acid methylation". Development. 104 (iv): 657–668. doi:10.1242/dev.104.4.657. PMID 3268408.
- ^ Giles KL (1971). "Dedifferentiation and Regeneration in Bryophytes: A Selective Review". New Zealand Journal of Botany. 9 (4): 689–94. doi:10.1080/0028825x.1971.10430231. Archived from the original on 2008-12-04. Retrieved 2008-01-01 .
- ^ Schnabel Thousand, Marlovits S, Eckhoff Chiliad, et al. (January 2002). "Dedifferentiation-associated changes in morphology and gene expression in main homo articular chondrocytes in cell culture". Osteoarthr. Cartil. 10 (1): 62–lxx. doi:10.1053/joca.2001.0482. PMID 11795984.
- ^ Sell S (December 1993). "Cellular origin of cancer: dedifferentiation or stalk cell maturation arrest?". Environ. Wellness Perspect. 101 (Suppl v): 15–26. doi:10.2307/3431838. JSTOR 3431838. PMC1519468. PMID 7516873.
- ^ Tsonis PA (April 2004). "Stem cells from differentiated cells". Mol. Interv. iv (two): 81–3. doi:10.1124/mi.iv.ii.4. PMID 15087480. Archived from the original on 2016-05-23. Retrieved 2010-12-26 .
- ^ Ben-Tabou de-Leon Southward, Davidson EH (2007). "Gene regulation: gene control network in development" (PDF). Annu Rev Biophys Biomol Struct. 36 (191): 191–212. doi:10.1146/annurev.biophys.35.040405.102002. PMID 17291181.
- ^ Newman, Stuart A. (2020). "Cell differentiation: what have we learned in 50 years?". Journal of Theoretical Biology. 485: 110031. arXiv:1907.09551. Bibcode:2020JThBi.48510031N. doi:ten.1016/j.jtbi.2019.110031. PMID 31568790.
- ^ Knisely, Karen; Gilbert, Scott F. (2009). Developmental Biological science (eighth ed.). Sunderland, Mass: Sinauer Associates. p. 147. ISBN978-0-87893-371-half dozen.
- ^ a b c d e f thou Rudel and Sommer; The evolution of developmental mechanisms. Developmental Biological science 264, 15-37, 2003 Rudel, D.; Sommer, R. J. (2003). "The evolution of developmental mechanisms". Developmental Biological science. 264 (1): fifteen–37. doi:10.1016/S0012-1606(03)00353-1. PMID 14623229.
- ^ a b Yamamoto Y and WR Jeffery; Central role for the lens in cave fish heart degeneration. Science 289 (5479), 631-633, 2000 Yamamoto, Y.; Jeffery, Due west. R. (2000). "Central Function for the Lens in Cave Fish Eye Degeneration". Science. 289 (5479): 631–633. Bibcode:2000Sci...289..631Y. doi:10.1126/science.289.5479.631. PMID 10915628.
- ^ Kirk MM, A Ransick, SE Mcrae, DL Kirk; The relationship betwixt cell size and jail cell fate in Volvox carteri. Journal of Cell Biology 123, 191-208, 1993 Kirk, M. Thou.; Ransick, A.; McRae, S. East.; Kirk, D. L. (1993). "The relationship between cell size and cell fate in Volvox carteri". Journal of Jail cell Biology. 123 (1): 191–208. doi:10.1083/jcb.123.i.191. PMC2119814. PMID 8408198.
- ^ Rabajante JF, Babierra AL (January 30, 2015). "Branching and oscillations in the epigenetic mural of cell-fate determination" (PDF). Progress in Biophysics and Molecular Biology. 117 (2–3): 240–9. doi:10.1016/j.pbiomolbio.2015.01.006. PMID 25641423.
- ^ a b Lister R; et al. (2011). "Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stalk cells". Nature. 471 (7336): 68–73. Bibcode:2011Natur.471...68L. doi:10.1038/nature09798. PMC3100360. PMID 21289626.
- ^ a b c d e f g h Christophersen NS, Helin 1000 (2010). "Epigenetic command of embryonic stem cell fate". J Exp Med. 207 (11): 2287–95. doi:10.1084/jem.20101438. PMC2964577. PMID 20975044.
- ^ a b Thomson, M; Liu, S. J.; Zou, 50. N.; Smith, Z; Meissner, A; Ramanathan, S (2011). "Pluripotency factors in embryonic stem cells regulate differentiation into germ layers". Jail cell. 145 (half-dozen): 875–89. doi:x.1016/j.cell.2011.05.017. PMC5603300. PMID 21663792.
- ^ Zhu, J.; et al. (2013). "Genome-wide chromatin country transitions associated with developmental and environmental cues". Cell. 152 (3): 642–654. doi:10.1016/j.jail cell.2012.12.033. PMC3563935. PMID 23333102.
- ^ a b c Guenther MG, Immature RA (2010). "Repressive Transcription" (PDF). Science. 329 (5988): 150–1. Bibcode:2010Sci...329..150G. doi:10.1126/science.1193995. PMC3006433. PMID 20616255.
- ^ a b c Meissner A (2010). "Epigenetic modifications in pluripotent and differentiated cells". Nat Biotechnol. 28 (10): 1079–88. doi:10.1038/nbt.1684. PMID 20944600. S2CID 205274850.
- ^ Flake Overview
- ^ Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean Thousand, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (Mar 2003). "The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation". Molecular Prison cell. 11 (3): 721–9. doi:10.1016/S1097-2765(03)00091-1. PMID 12667454.
- ^ Ng HH, Robert F, Young RA, Struhl Grand (Mar 2003). "Targeted recruitment of Set1 histone methylase by elongating Political leader Ii provides a localized marking and memory of recent transcriptional activity". Molecular Jail cell. 11 (three): 709–xix. doi:10.1016/S1097-2765(03)00092-3. PMID 12667453.
- ^ Bernstein Be, Kamal M, Lindblad-Toh 1000, Bekiranov S, Bailey DK, Huebert DJ, McMahon Southward, Karlsson EK, Kulbokas EJ, Gingeras TR, Schreiber SL, Lander ES (Jan 2005). "Genomic maps and comparative assay of histone modifications in human and mouse". Cell. 120 (two): 169–81. doi:10.1016/j.jail cell.2005.01.001. PMID 15680324. S2CID 7193829.
- ^ Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Höfer T, Rippe K (2012). "Genome-broad nucleosome positioning during embryonic stem prison cell evolution". Nat Struct Mol Biol. nineteen (11): 1185–92. doi:10.1038/nsmb.2419. PMID 23085715. S2CID 34509771.
- ^ a b Whyte, W. A.; Bilodeau, S; Orlando, D. A.; Hoke, H. A.; Frampton, Thousand. M.; Foster, C. T.; Cowley, Due south. Grand.; Immature, R. A. (2012). "Enhancer decommissioning by LSD1 during embryonic stalk cell differentiation". Nature. 482 (7384): 221–5. Bibcode:2012Natur.482..221W. doi:10.1038/nature10805. PMC4144424. PMID 22297846.
- ^ a b c d due east f Mohammad HP, Baylin SB (2010). "Linking cell signaling and the epigenetic machinery". Nat Biotechnol. 28 (10): 1033–eight. doi:10.1038/nbt1010-1033. PMID 20944593. S2CID 6911946.
- ^
- ^ Liu South; et al. (2006). "Hedgehog Signaling and Bmi-1 Regulate Self-renewal of Normal and Cancerous Human Mammary Stem Cells". Cancer Res. 66 (12): 6063–71. doi:x.1158/0008-5472.CAN-06-0054. PMC4386278. PMID 16778178.
- ^ Leung C; et al. (2004). "Bmi1 is essential for cerebellar evolution and is overexpressed in human medulloblastomas". Nature. 428 (6980): 337–41. Bibcode:2004Natur.428..337L. doi:10.1038/nature02385. PMID 15029199. S2CID 29965488.
- ^ Zencak D; et al. (2005). "Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation". J Neurosci. 25 (24): 5774–83. doi:ten.1523/JNEUROSCI.3452-04.2005. PMC6724881. PMID 15958744.
- ^ a b Engler, AJ; Sen, South; Sweeney, HL; Discher, DE (August 2006). "Matrix Elasticity Directs Stem Cell Lineage Specification". Cell. 126 (4): 677–689. doi:10.1016/j.cell.2006.06.044. PMID 16923388. S2CID 16109483.
- ^ Guo, Jun; Wang, Yuexiu; Sachs, Frederick; Meng, Fanjie (2014-12-09). "Actin stress in prison cell reprogramming". Proceedings of the National Academy of Sciences. 111 (49): E5252–E5261. Bibcode:2014PNAS..111E5252G. doi:10.1073/pnas.1411683111. ISSN 0027-8424. PMC4267376. PMID 25422450.
- ^ Guilak, Farshid; Cohen, Daniel M.; Estes, Bradley T.; Gimble, Jeffrey Thousand.; Liedtke, Wolfgang; Chen, Christopher South. (2009-07-02). "Control of Stem Cell Fate by Physical Interactions with the Extracellular Matrix". Cell Stem Cell. 5 (ane): 17–26. doi:x.1016/j.stem.2009.06.016. PMC2768283. PMID 19570510.
- ^ "Billion-twelvemonth-former fossil reveals missing link in the development of animals". phys.org . Retrieved nine May 2021.
- ^ "Billion-year-old fossil found preserved in Torridon rocks". BBC News. 2021-04-29. Retrieved 22 May 2021.
- ^ Strother, Paul Grand.; Brasier, Martin D.; Wacey, David; Timpe, Leslie; Saunders, Martin; Wellman, Charles H. (13 April 2021). "A possible billion-year-old holozoan with differentiated multicellularity". Current Biological science. 31 (12): 2658–2665.e2. doi:x.1016/j.cub.2021.03.051. ISSN 0960-9822. PMID 33852871.
 Available nether CC Past 4.0.
Available nether CC Past 4.0.
Source: https://en.wikipedia.org/wiki/Cellular_differentiation



0 Response to "What would happen to cell if they did not differentiate"
Post a Comment